Globin conservation analysis
The Globin family revisited
Last lab class we used HBB (beta globin) as a search query of the RefSeq database using PSI-BLAST. This allowed us to find distant human globin sequences such as neuroglobin, cytoglobin and of course myoglobin. Critically we could not find these more distant (i.e. diverged in sequence) globins with a regular BLAST search.
We also examined and compared the atomic resolution structures of beta globin and myoglobin and found them to be remarkably similar. A key findings here was that the traces of common ancestry are very much apparent at the structural level even when the sequences of these homologues have diverged beyond the detection limits of conventional methods like protein BLAST. Recall that we had to do quite a bit of extra work with PSI-BLAST to find these homologues sequences. If this does not sound familiar please go back to the lab section for class 3.
Here we use the results from PSI-BLAST (last lab session end of section 3) together with some R code to dig deeper into these results for mechanistic insight into important functional features of all globins.
Step 1. Read our globin alignment
We will use the bio3d R package to read the FASTA formatted alignment
we generated last day. Note that you will need to have the bio3d package
installed (via the R command install.packages("bio3d") for
this to work on your computer). We will talk more about R packages in a
couple of classes time.
library(bio3d)## Warning: package 'bio3d' was built under R version 4.1.2# Feel free to use your own alignment input here
inputfile <- "https://bioboot.github.io/bimm143_F21/class-material/globin_alignment.fa"
aln <- read.fasta( inputfile )Examine a print out of this alignment, note visually any conserved positions.
aln## 1 . . . . . 60
## beta ---------------MVHLTPEEKSAVTALWGKV--NVDEVGGEALGRLLVVYPWTQRFF
## delta ---------------MVHLTPEEKTAVNALWGKV--NVDAVGGEALGRLLVVYPWTQRFF
## gamma-2 ---------------MGHFTEEDKATITSLWGKV--NVEDAGGETLGRLLVVYPWTQRFF
## epsilon ---------------MVHFTAEEKAAVTSLWSKM--NVEEAGGEALGRLLVVYPWTQRFF
## gamma-1 ---------------MGHFTEEDKATITSLWGKV--NVEDAGGETLGRLLVVYPWTQRFF
## zeta ----------------MSLTKTERTIIVSMWAKISTQADTIGTETLERLFLSHPQTKTYF
## alpha ----------------MVLSPADKTNVKAAWGKVGAHAGEYGAEALERMFLSFPTTKTYF
## cytoglobin-X1 MEKVPGEMEIERRERSEELSEAERKAVQAMWARLYANCEDVGVAILVRFFVNFPSAKQYF
## mu -----------------MLSAQERAQIAQVWDLIAGHEAQFGAELLLRLFTVYPSTKVYF
## theta-1 ----------------MALSAEDRALVRALWKKLGSNVGVYTTEALERTFLAFPATKTYF
## cytoglobin MEKVPGEMEIERRERSEELSEAERKAVQAMWARLYANCEDVGVAILVRFFVNFPSAKQYF
## cytoglobin-X2 ------------------------------------------------------------
## myoglobin-1 ----------------MGLSDGEWQLVLNVWGKVEADIPGHGQEVLIRLFKGHPETLEKF
## neuroglobin ------------------MERPEPELIRQSWRAVSRSPLEHGTVLFARLFALEPDLLPLF
## myoglobin-2 ------------------------------------------------------------
##
## 1 . . . . . 60
##
## 61 . . . . . 120
## beta E-SFGDLSTPDAVM-GNPKVKAHGKKVLGAFSDGLAHLDNLKGT---FATLSELHCDKLH
## delta E-SFGDLSSPDAVM-GNPKVKAHGKKVLGAFSDGLAHLDNLKGT---FSQLSELHCDKLH
## gamma-2 D-SFGNLSSASAIM-GNPKVKAHGKKVLTSLGDAIKHLDDLKGT---FAQLSELHCDKLH
## epsilon D-SFGNLSSPSAIL-GNPKVKAHGKKVLTSFGDAIKNMDNLKPA---FAKLSELHCDKLH
## gamma-1 D-SFGNLSSASAIM-GNPKVKAHGKKVLTSLGDATKHLDDLKGT---FAQLSELHCDKLH
## zeta P-HF-------DLHPGSAQLRAHGSKVVAAVGDAVKSIDDIGGA---LSKLSELHAYILR
## alpha P-HF-------DLSHGSAQVKGHGKKVADALTNAVAHVDDMPNA---LSALSDLHAHKLR
## cytoglobin-X1 S-QFKHMEDPLEME-RSPQLRKHACRVMGALNTVVENLHDPDKVSSVLALVGKAHALKHK
## mu P-HL------SACQ-DATQLLSHGQRMLAAVGAAVQHVDNLRAA---LSPLADLHALVLR
## theta-1 S-H-------LDLSPGSSQVRAHGQKVADALSLAVERLDDLPHA---LSALSHLHACQLR
## cytoglobin S-QFKHMEDPLEME-RSPQLRKHACRVMGALNTVVENLHDPDKVSSVLALVGKAHALKHK
## cytoglobin-X2 ------MEDPLEME-RSPQLRKHACRVMGALNTVVENLHDPDKVSSVLALVGKAHALKHK
## myoglobin-1 D-KFKHLKSEDEMK-ASEDLKKHGATVLTALGGILKKKGHHEAE---IKPLAQSHATKHK
## neuroglobin QYNCRQFSSPEDCL-SSPEFLDHIRKVMLVIDAAVTNVEDLSSLEEYLASLGRKHRA-VG
## myoglobin-2 ------------MK-ASEDLKKHGATVLTALGGILKKKGHHEAE---IKPLAQSHATKHK
## * ^ ^ *
## 61 . . . . . 120
##
## 121 . . . . . 180
## beta VDPENFRLLGNVLVCVLAHHFGKEFTPPVQAAYQKVVAGVANALAHKYH-----------
## delta VDPENFRLLGNVLVCVLARNFGKEFTPQMQAAYQKVVAGVANALAHKYH-----------
## gamma-2 VDPENFKLLGNVLVTVLAIHFGKEFTPEVQASWQKMVTGVASALSSRYH-----------
## epsilon VDPENFKLLGNVMVIILATHFGKEFTPEVQAAWQKLVSAVAIALAHKYH-----------
## gamma-1 VDPENFKLLGNVLVTVLAIHFGKEFTPEVQASWQKMVTAVASALSSRYH-----------
## zeta VDPVNFKLLSHCLLVTLAARFPADFTAEAHAAWDKFLSVVSSVLTEKYR-----------
## alpha VDPVNFKLLSHCLLVTLAAHLPAEFTPAVHASLDKFLASVSTVLTSKYR-----------
## cytoglobin-X1 VEPVYFKILSGVILEVVAEEFASDFPPETQRAWAKLRGLIYSHVTAAYKEVGWVQQVPNA
## mu VDPANFPLLIQCFHVVLASHLQDEFTVQMQAAWDKFLTGVAVVLTEKYR-----------
## theta-1 VDPASFQLLGHCLLVTLARHYPGDFSPALQASLDKFLSHVISALVSEYR-----------
## cytoglobin VEPVYFKILSGVILEVVAEEFASDFPPETQRAWAKLRGLIYSHVTAAYKEVGWVQQVPNA
## cytoglobin-X2 VEPVYFKILSGVILEVVAEEFASDFPPETQRAWAKLRGLIYSHVTAAYKEVGWVQQVPNA
## myoglobin-1 IPVKYLEFISECIIQVLQSKHPGDFGADAQGAMNKALELFRKDMASNYKELGFQG-----
## neuroglobin VKLSSFSTVGESLLYMLEKCLGPAFTPATRAAWSQLYGAVVQAMSRGWDGE---------
## myoglobin-2 IPVKYLEFISECIIQVLQSKHPGDFGADAQGAMNKALELFRKDMASNYKELGFQG-----
## ^ ^ ^ * ^ ^
## 121 . . . . . 180
##
## 181 . . 205
## beta -------------------------
## delta -------------------------
## gamma-2 -------------------------
## epsilon -------------------------
## gamma-1 -------------------------
## zeta -------------------------
## alpha -------------------------
## cytoglobin-X1 TTHSSWRRSPEGSWGRQASCPSCC-
## mu -------------------------
## theta-1 -------------------------
## cytoglobin TTPP-------------ATLPSSGP
## cytoglobin-X2 TTHSSWRRSPEGSWGRQASCPSCC-
## myoglobin-1 -------------------------
## neuroglobin -------------------------
## myoglobin-2 -------------------------
##
## 181 . . 205
##
## Call:
## read.fasta(file = inputfile)
##
## Class:
## fasta
##
## Alignment dimensions:
## 15 sequence rows; 205 position columns (92 non-gap, 113 gap)
##
## + attr: id, ali, callDoing things visually and manually counting characters in an alignment such as this is not much fun and very susceptible to making mistakes. Let’s use some code to help.
Step 2. Identifying conserved positions
First we will score positional conservation in the alignment with the
conserv() function.
sim <- conserv(aln)Make a quick plot of alignment position vs conservation score.
plot(sim, typ="h", xlab="Alignment position", ylab="Conservation score")Here the high bars represent the most conserved amino-acid positions.
Let’s order our sim vector of conservation scores to put
the most conserved (highest scoring) first. What positions are the most
conserved?
inds <- order(sim, decreasing=TRUE)
head(sim[inds])## [1] 1.0000000 1.0000000 1.0000000 0.8933333 0.8933333 0.8666667The top three positions are absolutely invariant here. Let’s use the
ordering indices determened with the order() function above
to find out more about the positions.
To help we can make a dataframe that has all the information we might want, such as position number in the alignment, the amino acid in our first beta globin sequence and conservation score.
positions <- data.frame(pos=1:length(sim),
aa=aln$ali[1,],
score=sim)
head(positions)## pos aa score
## 1 1 - 0.00952381
## 2 2 - 0.00952381
## 3 3 - 0.00952381
## 4 4 - 0.00952381
## 5 5 - 0.00952381
## 6 6 - 0.00952381Note the first 6 positions of the alignment are gap characters in the beta globin sequence and have very low (close to zero) conservation scores.
Let’s use the ordering vector from above to look at the most conserved positions:
head( positions[inds,] )## pos aa score
## 83 83 H 1.0000000
## 115 115 H 1.0000000
## 145 145 F 1.0000000
## 87 87 V 0.8933333
## 155 155 K 0.8933333
## 168 168 Y 0.8666667Nice, this shows us that positions 83, 115 and 145 are indeed very conserved.
Let’s convert the one-letter code for these amino acids to 3 letter code.
aa123(positions[inds,]$aa)[1:3]## [1] "HIS" "HIS" "PHE"In the figure below I mapped these top two Histidne amino acids to atomic structure (we will see how to do this in R later in the course). Note that these conserved positions are responsible for coordinating the all important iron containing heam group (a.k.a. Porphyrin ring).
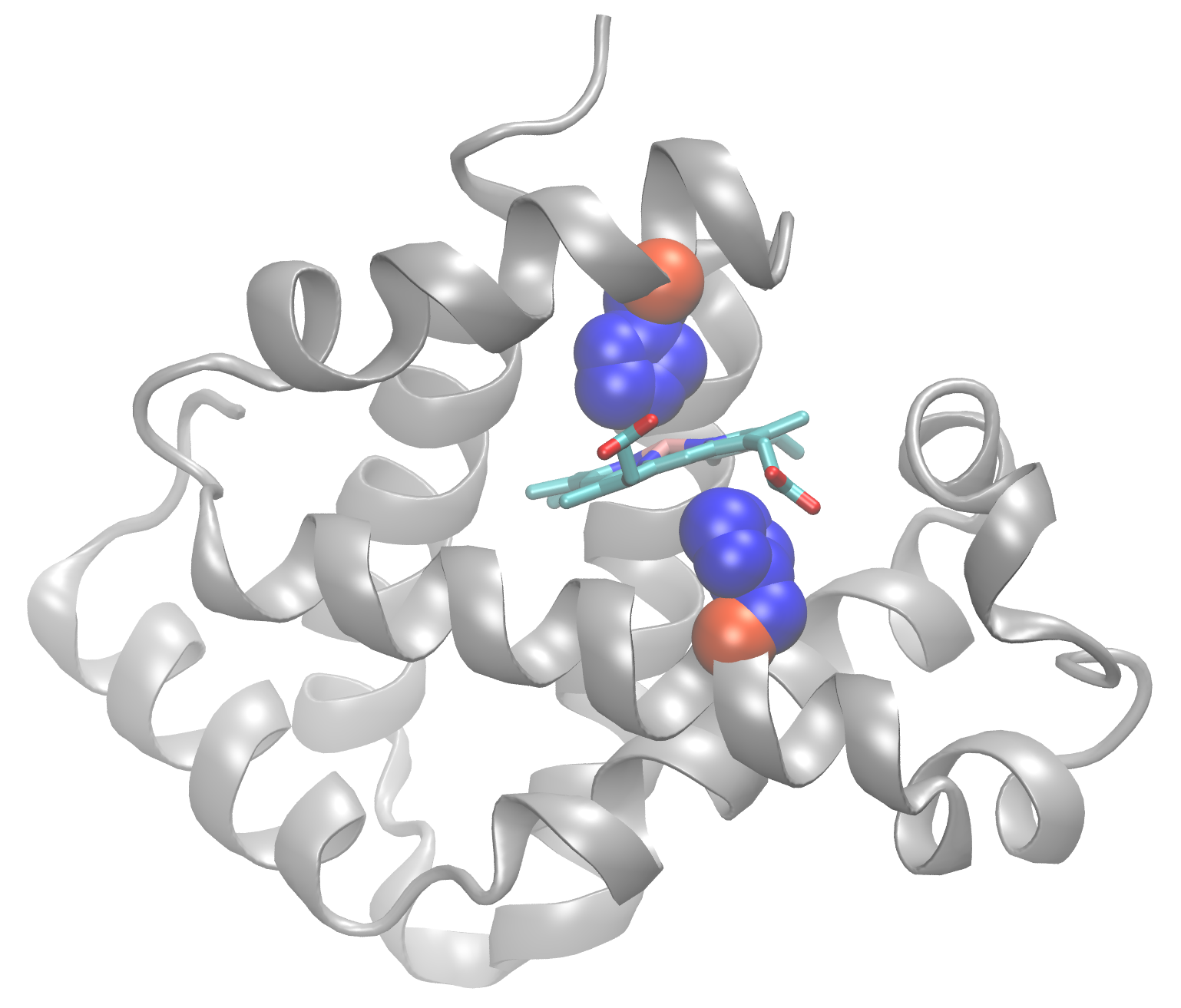
Beta globin structure with two conserved Histidine resides (blue spheres) and Porphyrin ring (cyan) shown.
Step 3. Relationship between sequences
Here we calculate the sequence identity between all pairs of
sequences (with the seqidentity() function) and use the
resulting matrix of identity scores to generate a heatmap visualization
of the relationship between sequences. This is a common visualization in
bioinformatcs and we will revisit it several times in the course
(especially with NGS data analysis).
Note here we use the pheatmap package, which again needs
to be installed on your computer with the R code
install.packages("pheatmap").
library(pheatmap)
ide <- seqidentity(aln)
pheatmap((1-ide))